Experts in metatranscriptomics
Unveiling the living fraction of organisms in a complex sample with our metatranscriptomics service
AllGenetics' metatranscriptomics service provides complete information about the living fraction of organisms in a complex sample. This technique takes advantage of the fact that RNA rapidly degrades in dead individuals. Therefore, it can help elucidate the genes that are being actively transcribed in a certain environment. Metatranscriptomics yields extensive information about functional diversity and operating metabolic pathways and networks.
We can apply metagenomics and metatranscriptomics to a variety of fields, such as microbial ecology, soil biology, ecosystem monitoring, and host-microbe interactions. Remarkably, the metatranscriptomic approach is able to detect ssRNA and dsRNA viruses.
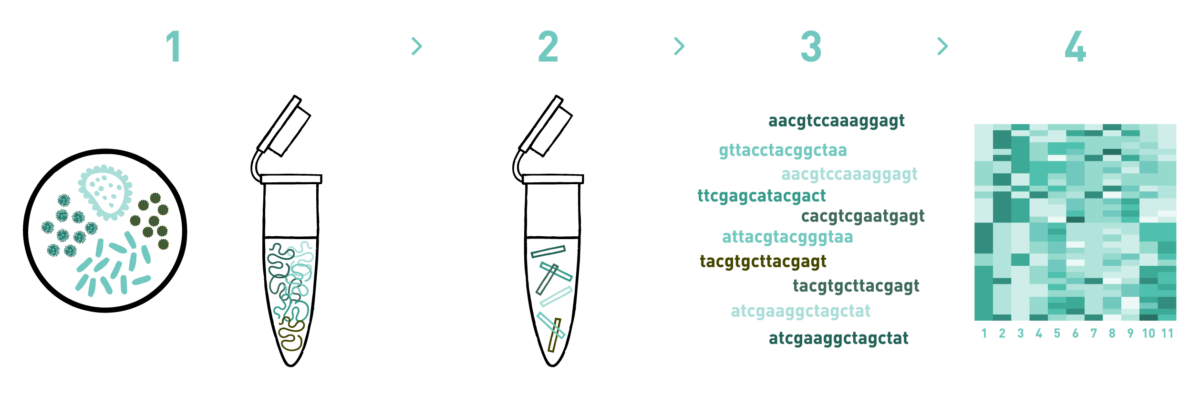
Step 1
We isolate RNA from different types of samples such as soil, water, biofilm, sludge, etc.
Step 2
We retrotranscribe the RNA into cDNA and prepare shotgun libraries for all of the biodiversity present in the sample.
Step 3
We sequence the libraries using the Illumina MiSeq or NovaSeq technologies. We can adapt the output to your specific requirements.
Step 4
We analyse the high-throughput sequencing data to obtain your annotated metagenomes.
For your convenience, we can carry out the entire project or only those steps you require. As a norm, steps 2 and 3 should be ordered jointly.