Experts in snp genotyping
Get to know our SNP genotyping service
AllGenetics' SNP genotyping service takes advantage of the new genotyping-by-sequencing method MobiSeq. MobiSeq was developed by Dr. Anders Johannes Hansen's group at the University of Copenhagen. It generates genomic data based on the PCR-enrichment of repetitive elements scattered throughout the genome. The way the technique works is by designing a primer at the conserved end of a repetitive element and sequencing its flanking region outside the repetitive element. Thousands of such flanking regions are sequenced in each individual, allowing for SNP identification and genotyping across all the individuals being studied.
Despite the need for genomic information on the species under study, MobiSeq has proven advantages over other genotyping-by-sequencing methods. For example, MobiSeq:
- Is species-specific, so DNA from symbiont species will not interfere in the SNP genotyping process.
- Is suitable for low quality and low concentration DNA samples.
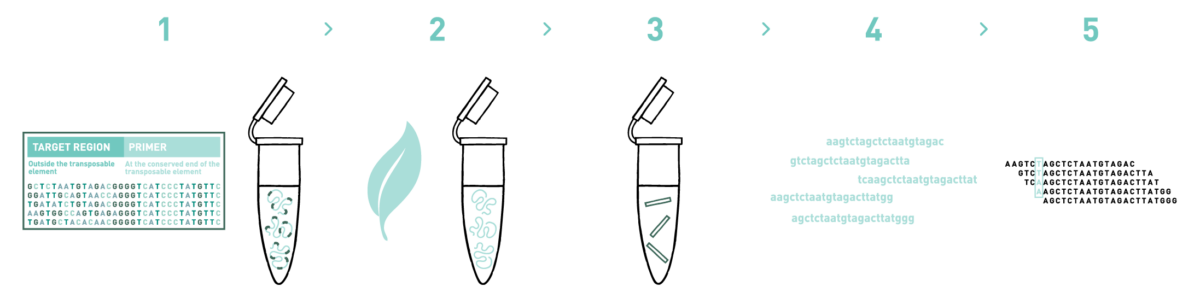
Step 1
We sequence one individual of the target species in the Illumina NovaSeq platform to obtain 90 gigabases of genomic information that will be used for primer design. Then, we identify a suitable repetitive element and design a primer within a conserved region in its 5' or 3' end.
Step 2
We isolate DNA from your samples. We have several optimised DNA isolation protocols, depending on the starting biological material.
Step 3
We prepare the reduced-representation libraries following the MobiSeq protocol.
Step 4
We sequence the libraries in the Illumina NovaSeq platform. Up to hundreds of libraries can be multiplexed in the same sequencing run and enough coverage of the loci under study can still be obtained.
Step 5
We analyse the high-throughput sequencing data to obtain the genotypes of the individuals under study.
For your convenience, we can carry out the entire project or only those steps you require. As a norm, steps 3 and 4 should be ordered jointly.