Experts in identifying food ingredients by DNA metabarcoding
Infer the complete list of ingredients in mixed food products
Thanks to high-throughput sequencing and DNA metabarcoding we are able to simultaneously identify the multiple species present in a complex food matrix, even without a prior knowledge of the expected composition. This is what makes our food ingredient identification service a powerful tool in a number of applications, including:
- Identification of constituent plant species in herbal products such as tea blends, spices, plant-based medicines, and food supplements.
- Characterisation of the floral composition of honey.
- Detection of oil adulteration.
- Species verification of fruit-based products and vegetal beverages (juices, jams, yoghurt).
- Identification of animal species in processed meat products.
- Microbiome profiling of dairy products.
- Characterisation of wine microorganisms.
Importantly, DNA metabarcoding can be applied to processed material containing highly degraded DNA, such as herbal extracts, oils, powders, or capsules.
Our DNA metabarcoding-based analyses can test the homogeneity of a sample, check if a species of interest is present in a complex mixture, or infer the complete list of ingredients in a sample. Therefore, systematic analysis of batches of products can prevent harmful species substitution and detect unlisted species being used as fillers, thereby ensuring consumer’s confidence and brand protection.
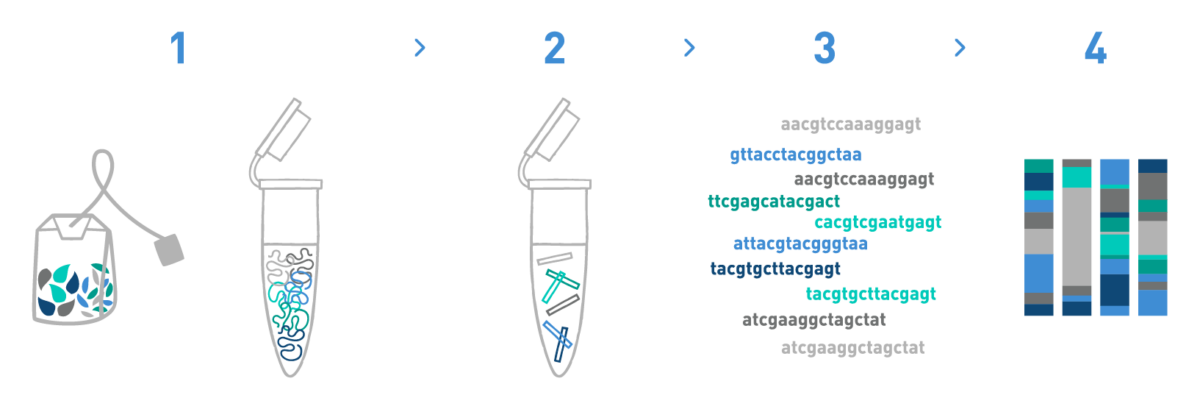
Step 1
We isolate DNA from different food matrices such as herbal blends, juices, dairy products, etc.
Step 2
We prepare DNA metabarcoding libraries for the target taxonomic group(s) -bacteria, fungi, algae, plants or animals-. For this, we use universal or in-house-developed primer pairs.
Step 3
We sequence the DNA metabarcoding libraries using Illumina's MiSeq or NovaSeq technologies.
Step 4
We analyse the data obtained in the previous step to obtain the taxonomic composition of the samples.
What you receive
- A list of the Operational Taxonomic Units (OTUs) retrieved per sample.
- The number of sequence reads assigned to each OTU in each sample.
- A graphic representation of the relative abundances of the different OTUs in each sample.
What we need
Your samples appropriately preserved depending on the sample type (ethanol, silica gel, frozen, or another suitable preservation method). If required, we provide sampling kits and sampling collection guidelines to ensure that your samples arrive at our lab in optimal conditions.