Experts in genome-wide association studies
Improve your trait selection method for plant breeding
Marker assisted selection (MAS) allows the breeder to obtain results in a more efficient way compared to conventional selection methods. Among the various approaches available, selection by detection of candidate genes or markers through association studies (genome-wide association studies, GWAS) is one of the most widely used. GWAS has the advantage over other methods that it does not require targeted crosses, which for some production species is crucial because it saves much time. Among the variety of GWAS approaches applied in breeding programs, AllGenetics can offer the following:
Case-control association testing
Genetic association testing with quantitative trait, including:
- Genome wide scanning or genome wide association mapping.
- General linear model (GLM).
- Mixed linear model (MLM).
- Single trait GWAS (stGWAS).
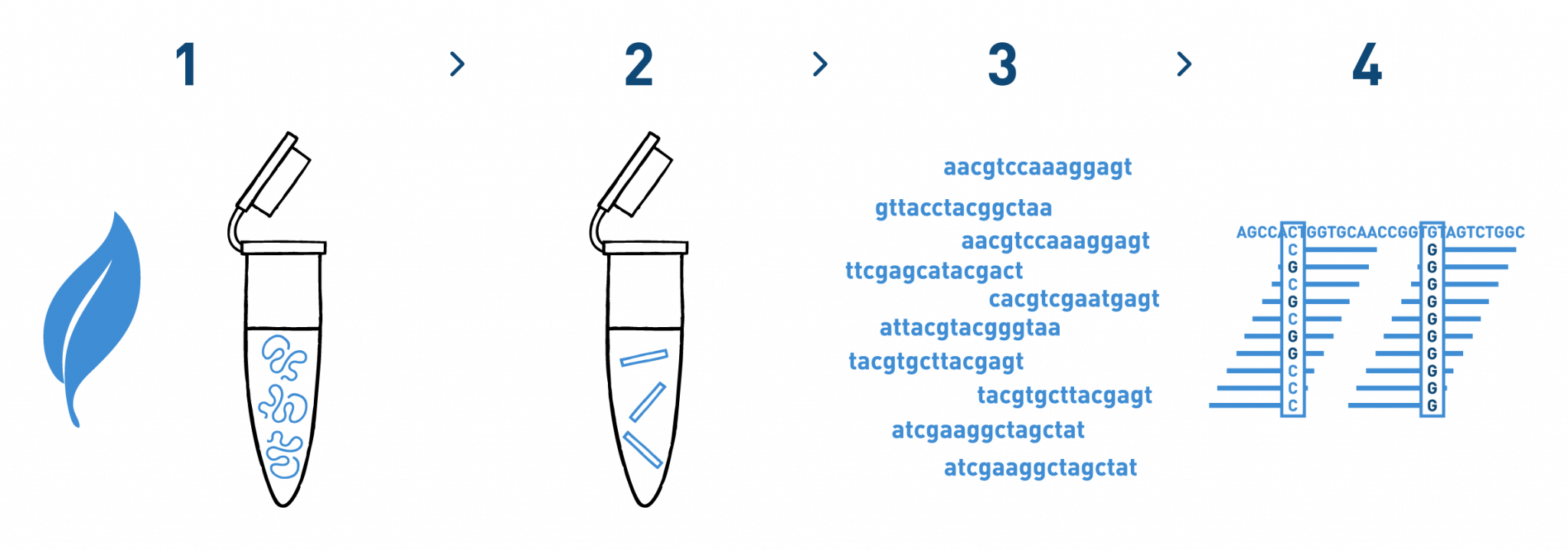
Step 1
We isolate DNA from your samples. We have adapted different DNA isolation protocols, depending on the starting biological material.
Step 2
We prepare genomic libraries using the kits recommended by Illumina.
Step 3
We sequence the libraries using the Illumina NovaSeq technology.
Step 4
We analyse the high-throughput sequencing data to identify and genotype the selected SNPs in each of the samples analysed. Then we proceed with the genome-wide association studies.
What you receive
- A table with the alleles called at each locus in each individual.
- A list of gene-associated SNPs.
- The results of genotype likelihood tests for diversity estimations.
- The preliminary quality control analyses for MAF, population structure (Bayesian approaches and principal component analysis - PCA), kinship, and linkage disequilibrium.
- The GWAS interpretation results include p-value, R2 value, QQ-plots, Manhattan plots, and GWAS power.
- Post GWAS (pGWAS) analyses for Identification of candidate genes, primer design for MTAs, and genotyping in populations of interest.