Experts in identifying grapevine genes of interest by RNA-seq
Get to know how we analyse grapevine expressed genes by RNA-seq
RNA-seq is a powerful, cost-effective tool for surveying plant response to specific conditions. Knowing which genes are being expressed under certain circumstances can help you select grapevine varieties or cultivars more resistant to stressors such as drought, frost, or pathogens.
At AllGenetics we use high-throughput sequencing to identify candidate genes underlying phenotypic traits of interest, providing invaluable information for genetic improvement programmes.
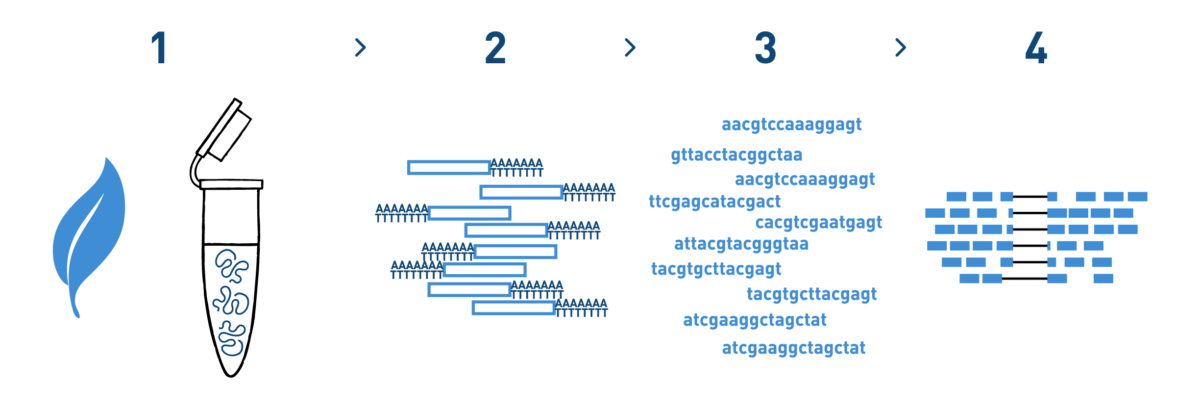
Step 1
We isolate RNA from your grapevine samples.
Step 2
We purify the RNA fraction you are interested in (total RNA, mRNA, small RNAs...) and prepare cDNA libraries using the kits recommended by Illumina.
Step 3
We sequence the libraries using the Illumina NovaSeq technology. Millions of reads per library are obtained in this step.
Step 4
We analyse the high-throughput sequencing data obtained in the previous step.
What you receive
- Gene expression quantification.
- Detection of differential gene expression between conditions.
What we need
Your plant tissue samples which have been appropriately preserved for RNA isolation. We provide sampling kits and sampling collection guidelines to ensure that your samples arrive at our lab in optimal conditions.