Experts in DNA metabarcoding
Diversity analysis of environmental samples with our DNA metabarcoding service
With our DNA metabarcoding service we can identify organisms down to various taxonomic levels and compare the diversity among samples. DNA metabarcoding can be used even when DNA is degraded. Therefore, it is possible to analyse species diversity in samples such as soil, faeces, or sediments.
AllGenetics' expertise on DNA metabarcoding encompasses DNA extraction from difficult matrices, custom primer design, or the use of blocking primers to prevent amplification of host DNA. Even though we run most of our projects in Illumina's short read sequencers (MiSeq and NovaSeq), we use Oxford Nanopore's and PacBio's long-read technologies when it becomes necessary to obtain longer barcodes. That way we make sure we get enough taxonomic resolution.
Our bioinformaticians will tailor our pipelines and databases to the specifics of your research project to obtain sound biodiversity estimates from your samples.
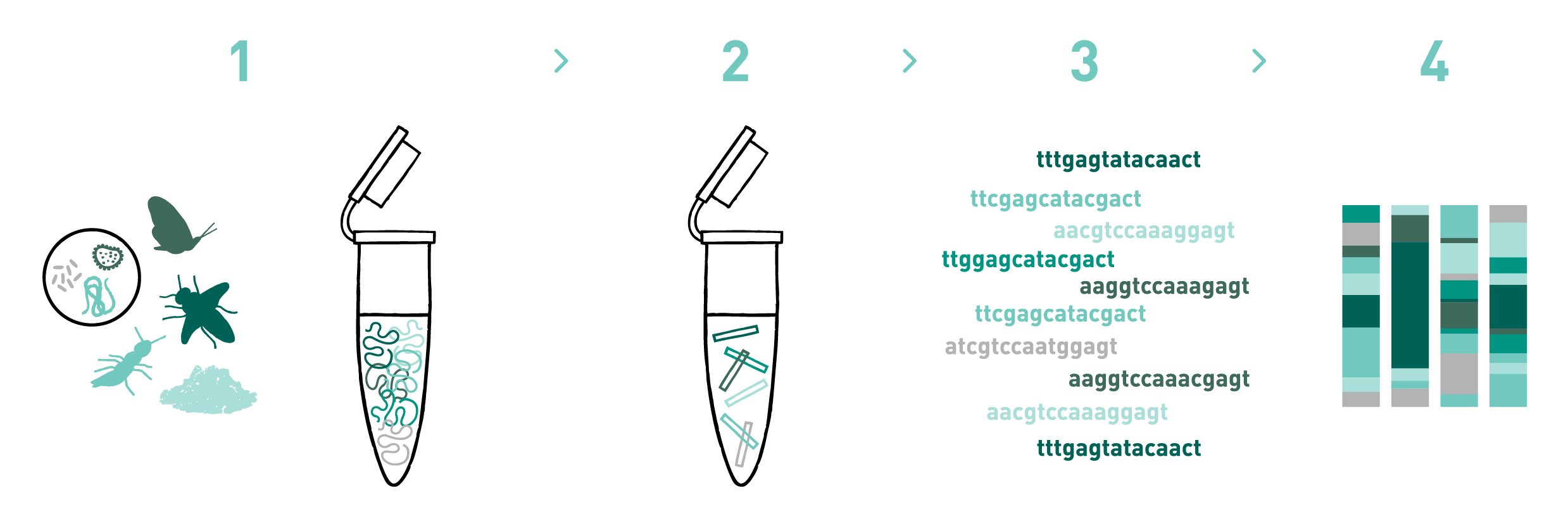
Step 1
We isolate DNA from different materials such as soil, water, faeces, sediments, organic residues, pollen, etc.
Step 2
We prepare DNA metabarcoding libraries for the target taxonomic group(s) -bacteria, fungi, plants, protists, and/or metazoans-. To do so we use universal or in-house-developed primer pairs. We can also design and use blocking primers during library preparation, if needed.
Step 3
We sequence the DNA metabarcoding libraries using Illumina's MiSeq or NovaSeq technologies.
Step 4
We analyse the data obtained in the previous step to obtain the taxonomic composition of the samples.
For your convenience, we can carry out the entire project or only those steps you require. As a norm, steps 2 and 3 should be ordered jointly.