Experts in monitoring microbial communities in bioreactors by DNA metabarcoding
An effective tool to track the evolution of microbial communities in industrial processes
The efficiency of aerobic or anaerobic digestion processes can change over time. This change is associated to shifts in the composition of microbial communities in bioreactors. DNA metabarcoding is the tool of choice to monitor changes in microbial communities over time, so that they can be associated to changes in the efficiency of the digestion process. In fact, understanding complex microbial communities and their activities is essential for the successful exploitation of bioenergy and waste treatment facilities.
At AllGenetics, we use DNA metabarcoding to obtain diversity data for various taxonomic groups (bacteria, archaea, algae) from a wide variety of bioprocesses like biogas production through anaerobic digestion or waste water treatment. By using DNA metabarcoding, it is possible to identify microbial indicators of optimal performance, as well as to establish warning indicators of potential process failure.
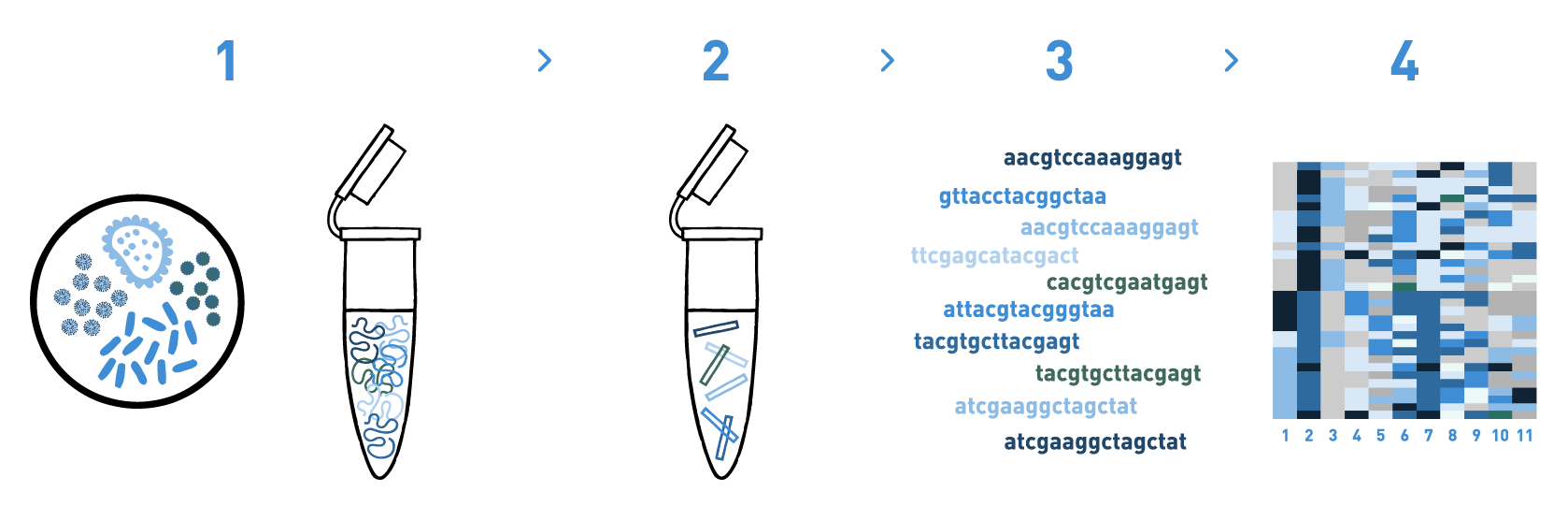
Step 1
We isolate DNA from the bioreactor samples.
Step 2
We prepare DNA metabarcoding libraries for the target taxonomic group(s) -bacteria, fungi, or algae-. For this, we use universal or in-house-developed primer pairs.
Step 3
We sequence the DNA metabarcoding libraries using Illumina's MiSeq or NovaSeq technologies.
Step 4
We analyse the data obtained in the previous step to obtain the taxonomic composition of the samples.
What you receive
- A list of the Operational Taxonomic Units (OTUs) retrieved per sample.
- The number of sequence reads assigned to each OTU in each sample.
- A graphic representation of the relative abundances of the different OTUs in each sample.
What we need
Your samples appropriately preserved depending on the sample type (ethanol, frozen, or another suitable preservation method). If required, we provide sampling kits and sampling collection guidelines to ensure that your samples arrive at our lab in optimal conditions.